

Welcome to the Cardiac MRI Reconstruction Challenge 2024 (CMRxRecon2024)!
The CMRxRecon2024 (also known as CMRxUniversalRecon) Challenge is a part of the 27th International Conference on Medical Image Computing and Computer Assisted Intervention, MICCAI 2024, which will be held from October 6th to 10th 2024 in Marrakesh, Morocco.
Learn moreThe objective of establishing the CMRxUniversalRecon challenge (Toward Universal Reconstruction) is to provide a benchmark that enables the broader research community to contribute to the important work of accelerated CMR imaging with universal approaches that allow more diverse applications and better performance in real-world deployment in various environments.
Cardiac magnetic resonance imaging (CMR) has emerged as a
crucial imaging technique for diagnosing cardiac diseases, thanks to its
excellent soft tissue contrast and non-invasive nature. However, a notable
limitation of MRI is its slow imaging speed, which causes patient discomfort
and introduces motion artifacts into the images.
To accelerate image acquisition, CMR image reconstruction
(recovering high-quality clinical interpretable images from highly
under-sampled k-space data) has gained significant attention in recent years. Particularly,
AI-based image reconstruction algorithms have shown great potential in
improving imaging performance by utilizing highly under-sampled data. Currently, the field of CMR
reconstruction lacks publicly available, standardized, and high-quality datasets for the development and
assessment for AI-based CMR reconstruction. In the first run of the 'CMRxRecon' challenge (MICCAI 2023), we
have provided cine and mapping data from a total of 300 subjects and the technical infrastructure as well as
a baseline model for CMR reconstructions. The results of 'CMRxRecon' 2023 demonstrated that deep learning
methods demonstrated significantly superior performance compared to traditional methods such as SENSE and
GRAPPA in a single task scenario.
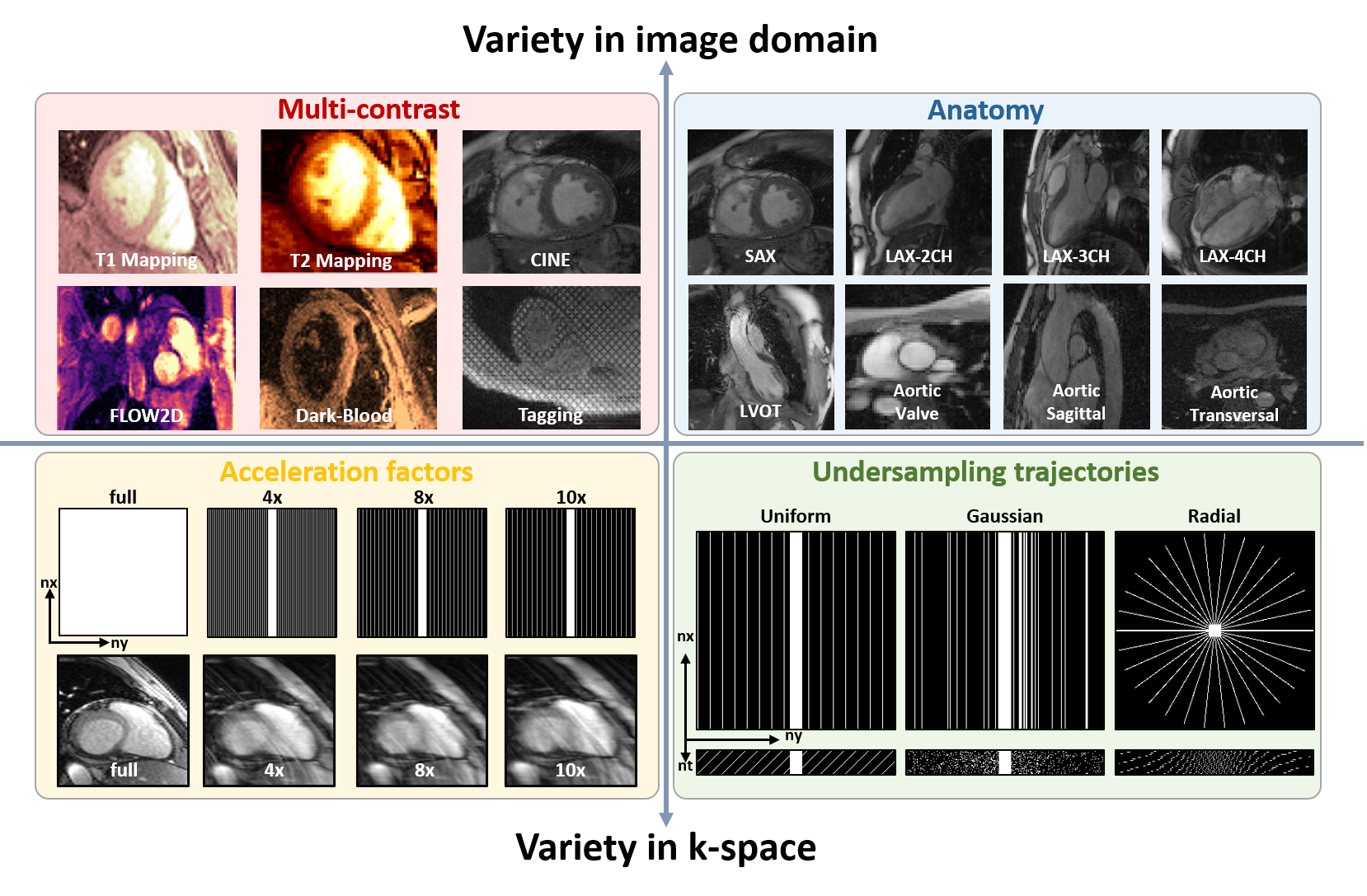
As we all know, CMR imaging has the nature of multi-contrast, e.g., cardiac cine, mapping, tagging,
phase-contrast, and dark-blood imaging. It also includes imaging of different anatomical views such as
long-axis (2-chamber, 3-chamber, and 4-chamber), short-axis, outflow tract, and aortic (cross-sectional and
sagittal views). Additionally, accelerated imaging trajectories, including uniformly undersampling and
variable-density sampling, are employed. Unfortunately,
conventional CNN-based reconstruction models often require training and deployment for each specific
imaging scenario (imaging sequence, view, and device vendor), limiting their clinical application in the
real world.
Thus, in this second run of the CMR reconstruction challenge we aim to make an important step towards
clinical implementation by extending the challenge scope in two directions:
1)Trustworthy reconstruction on multi-contrast CMR imaging
(two will be unseen in the training dataset) using a universal pre-trained reconstruction
model;
2)Robust reconstruction with diverse k-space trajectory and
various acceleration factors using a universal model.
The CMRxRecon2024 challenge includes two independent tasks. Each team can choose to participate one of them or both:
TASK 1: Multi-contrast CMR reconstruction
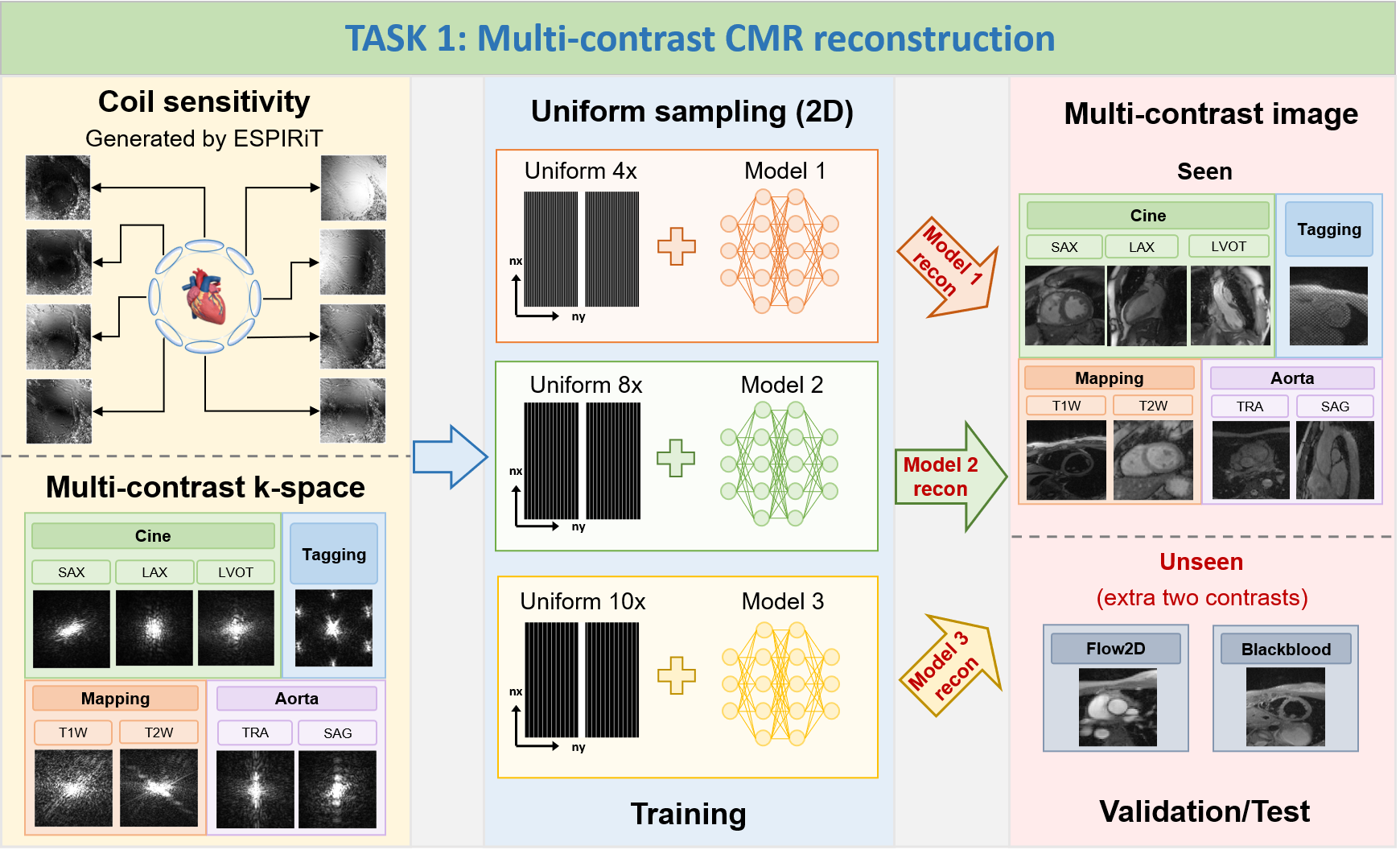
1) Goal: To
develop a contrast-universal model that can 1) provide
high-quality image reconstruction for highly-accelerated uniform undersampling (acceleration factors are 4x,
8x and 10x, ACS not included for calculations); 2) being
able to process multiple contrast reconstructions with different sequences, views, and scanning protocols
using a single universal model. The proposed method is supposed to offer a unified framework that can handle
various imaging contrasts, allowing for faster and more robust reconstructions across different CMR
protocols.
2) Note: In TASK 1, participants are allowed
to train three individual contrast-universal models to
respectively reconstruct multi-contrast data at the
aforementioned three acceleration factors; TrainingSet
includes Cine, Aorta, Mapping, and Tagging;
ValidationSet and TestSet include Cine, Aorta, Mapping,
Tagging, and other two unseen contrasts (Flow2d and
BlackBlood); the data size of Cine, Aorta, Mapping, Tagging, and Flow2d is 5D (nx,ny,nc,nz,nt); the
data
size of BlackBlood is 4D (nx,ny,nc,nz); the size of all
undersampling masks is 2D (nx,ny), the central 16
lines (ny) are always fully sampled to be used as autocalibration signals (ACS).
TASK 2: Random sampling CMR reconstruction
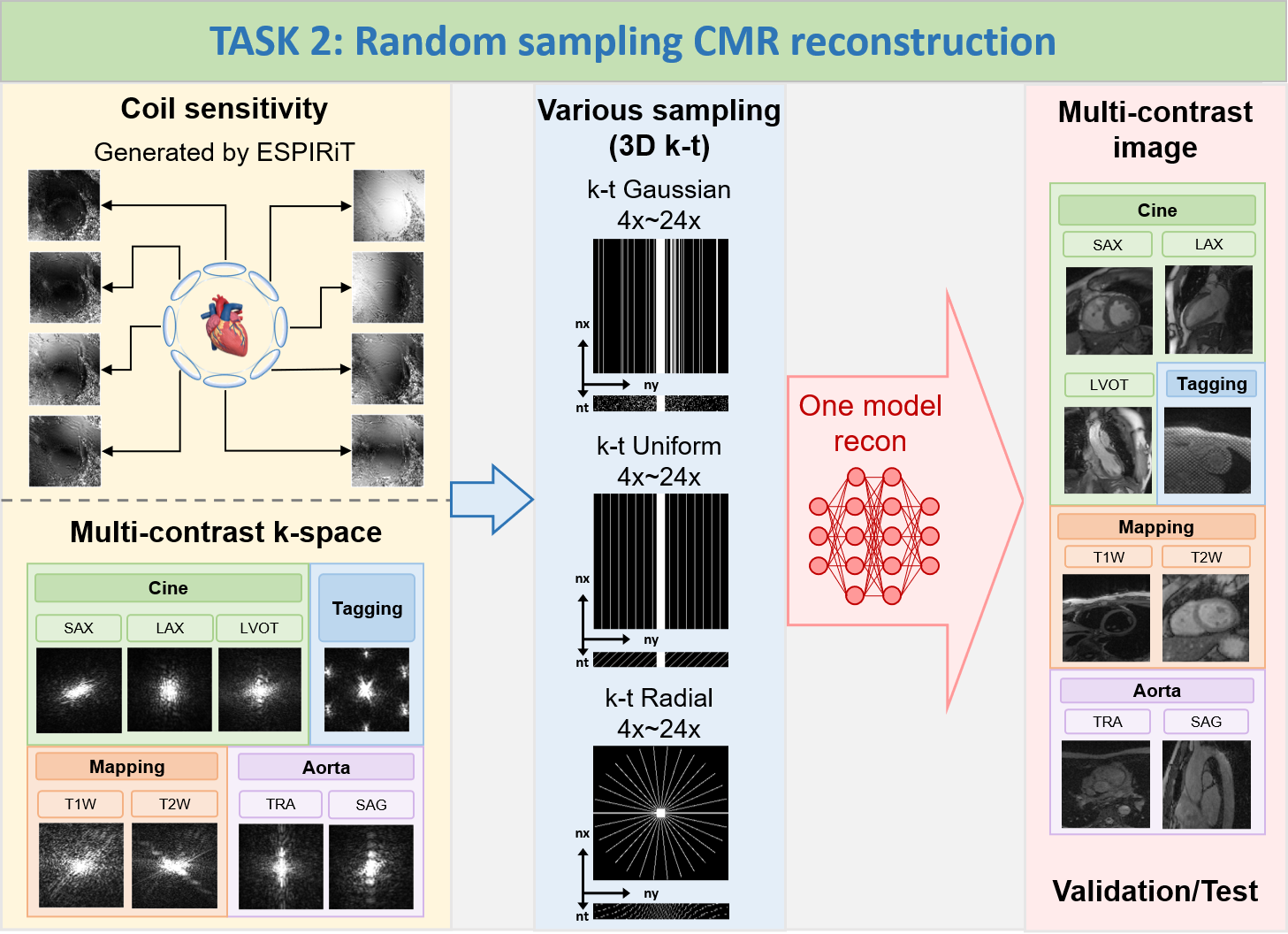
1) Goal: To
develop a sampling-universal model that can robustly reconstruct CMR images 1) from different k-space
trajectories (uniform, Guassian, and pseudo radial undersampling with temporal/parametric interleaving);
2)
at different acceleration factors (acceleration factors from 4x to 24x, ACS not included for calculations).
The proposed method is supposed to leverage deep learning algorithms to exploit the potential of random
sampling, enabling faster acquisition times while maintaining high-quality image reconstructions.
2) Note: In TASK 2, participants are allowed
to train only one universal model to reconstruct
various data at the different undersampling scenarios (including different k-space trajectories: uniform,
Guassian, and pseudo radial undersampling with temporal/parametric interleaving; and different acceleration
factors: 4x, 8x, 12x, 16x, 20x, 24x, ACS not included for calculations); TrainingSet includes Cine, Aorta,
Mapping, and Tagging; ValidationSet and TestSet also include Cine, Aorta, Mapping, and Tagging; the
data
size of Cine, Aorta, Mapping, and Tagging is 5D (nx,ny,nc,nz,nt); the size of all undersampling masks is 3D
(nx,ny,nt), the central 16 lines (ny, in ktUniform and ktGaussian) or central 16x16 regions (nx*ny,
in
ktRadial) are always fully sampled to be used as autocalibration signals (ACS).
We will provide monetary awards for the top 5 winners of each
task. The prize pool is exclusively sponsored by Philips.
Task 1: Multi-contrast CMR reconstruction
| Task | Winner | Monetary Awards | Certificate | Oral Presentation | Summary Paper Involved |
| Task 1 | Top 1 | $1,000 |  |
 |
 |
| Task 1 | Top 2 | $500 |  |
 |
 |
| Task 1 | Top 3 | $300 |  |
 |
 |
| Task 1 | Top 4 | $200 |  |
||
| Task 1 | Top 5 | $100 |  |
Task 2: Random sampling CMR reconstruction
| Task | Winner | Monetary Awards | Certificate | Oral Presentation | Summary Paper Involved |
| Task 2 | Top 1 | $1,000 |  |
 |
 |
| Task 2 | Top 2 | $500 |  |
 |
 |
| Task 2 | Top 3 | $300 |  |
 |
 |
| Task 2 | Top 4 | $200 |  |
||
| Task 2 | Top 5 | $100 |  |
All submissions will be reported in the leaderboard. Each participating team can participate in both
tasks. However, we only present the higher reward among the two
tasks to each team.
Prize-winning methods will be announced publicly as part of a scientific session at the MICCAI annual
meeting.
A total of 330 healthy volunteers are recruited for multi-contrast
CMR imaging in our imaging center. The dataset include
multi-contrast k-space data, consist of cardiac cine, T1/T2mapping, tagging, phase-contrast (i.e.,
flow2d), and dark-blood imaging. It also includes imaging of different anatomical views like long-axis
(2-chamber, 3-chamber, and 4-chamber), short-axis (SAX), left ventricul outflow tract (LVOT), and aorta
(transversal and sagittal views).
The released dataset includes 200 training data, 60
validation data and 70 test data.
Training cases including fully sampled k-space data will be provided in '.mat' format.
Validation cases include under-sampled k-space data, sampling trajectories, and autocalibration signals
(ACS, 16 lines or 16x16 regions) with various acceleration factors in '.mat' format.
Test cases include fully sampled k-space data, undersampled k-space data, sampling trajectories, and
autocalibration signals (ACS, 16 lines or 16x16 regions). Test cases will not be released before the
challenge ends.
1) Scanner:
Siemens 3T MRI scanner (MAGNETOM Vida)
2) Image acquisition: We follow the recommendations
of CMR exams reported in the previous publication (doi: 10.1007/s43657-02100018x,
10.1007/s43657-021-00018-x).
3) Dataset overview: The dataset will include
multi-contrast k-space data, consisting of cardiac cine, T1/T2 mapping, tagging, phase-contrast (i.e.,
flow2d), and dark-blood imaging. It also includes imaging of different anatomical views like long-axis (LAX,
including 2-chamber, 3-chamber, and 4-chamber), short-axis (SAX), left ventricul aroutflow tract (LVOT), and
aortic (transversal and sagittal views).
4) Scan protocol: We use 'TrueFISP' sequence for cine,
phase-constrast (i.e., flow2d), and tagging, and 'FLASH' sequence for T1/T2 mapping and dark-blood imaging.
For T1/T2 mapping, signals are collected at the end of the diastole with ECG triggering. Typically, 5~15
slices are acquired for each contrast. The cardiac cycle is segmented into 12~25 phases with a temporal
resolution of around 50 ms. Typical geometrical parameters include: spatial resolution 1.5×1.5 mm2, slice
thickness 8.0 mm, and slice gap 4.0 mm.
5) Pre-processing:The raw k-space data exported from the
scanner will be processed and transformed to '.mat' format using the script provided by our vendor. A readme
file will be provided to describe the content and usage of the data.
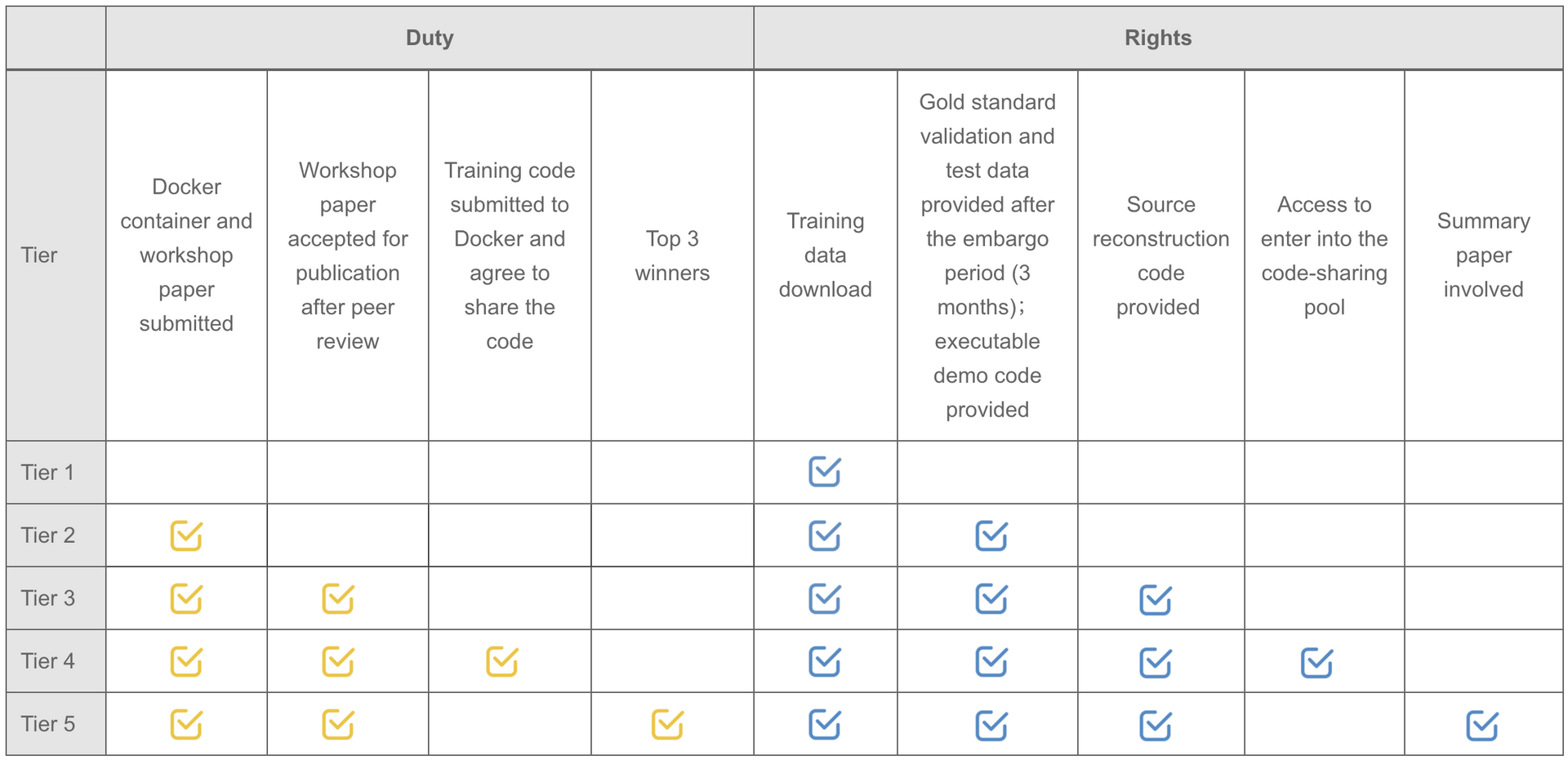
Note: Participants are not required to upload the complete training code. But teams willing to upload the original training code will be automatically entered into the code-sharing pool.
The schedule of the challenge is as follows. All deadlines are Pacific Standard Time (PST +0:00).
| 20 - Apr | Website opens for registration |
| 26 - Apr | Release training and validation data |
| 10 - May | Submission system opens for validation |
| 24 - Jun | Deadline for conference paper placeholder submission |
| 01 - Aug | Submission system opens for testing |
| 15 - Aug | Deadline for conference paper submission |
| 10 - Sept | Docker submission (test phase) deadline |
| 10 - Oct | Release final results |
| 10 - Nov | Proceeding ready paper release |
created with
Website Builder Software .
