CMR imaging is time-consuming due to the acquisition of a
large amount of raw data in the k-space (raw signal measurement). To address
this limitation, accelerated imaging techniques have been developed, which rely
on sub-Nyquist undersampling the k-space data. Random (incoherent) undersampling
of the k-space trajectory is one such approach that offers acceleration and
potential for improved image quality. The need for a universal model therefore arises
from the diverse sampling schemes of cardiac image acquisition: Different CMR
protocols often require distinct k-space sampling trajectories and acceleration
factors. Traditional reconstruction methods struggle to adapt to
these
variations, as they typically rely on sampling-specific algorithms. A universal
model offers a unified framework that can handle different sampling
trajectories and acceleration factors, providing flexibility and efficiency in
the reconstruction process. This approach is necessary to achieve consistent
and reliable reconstructions across diverse CMR imaging scenarios.
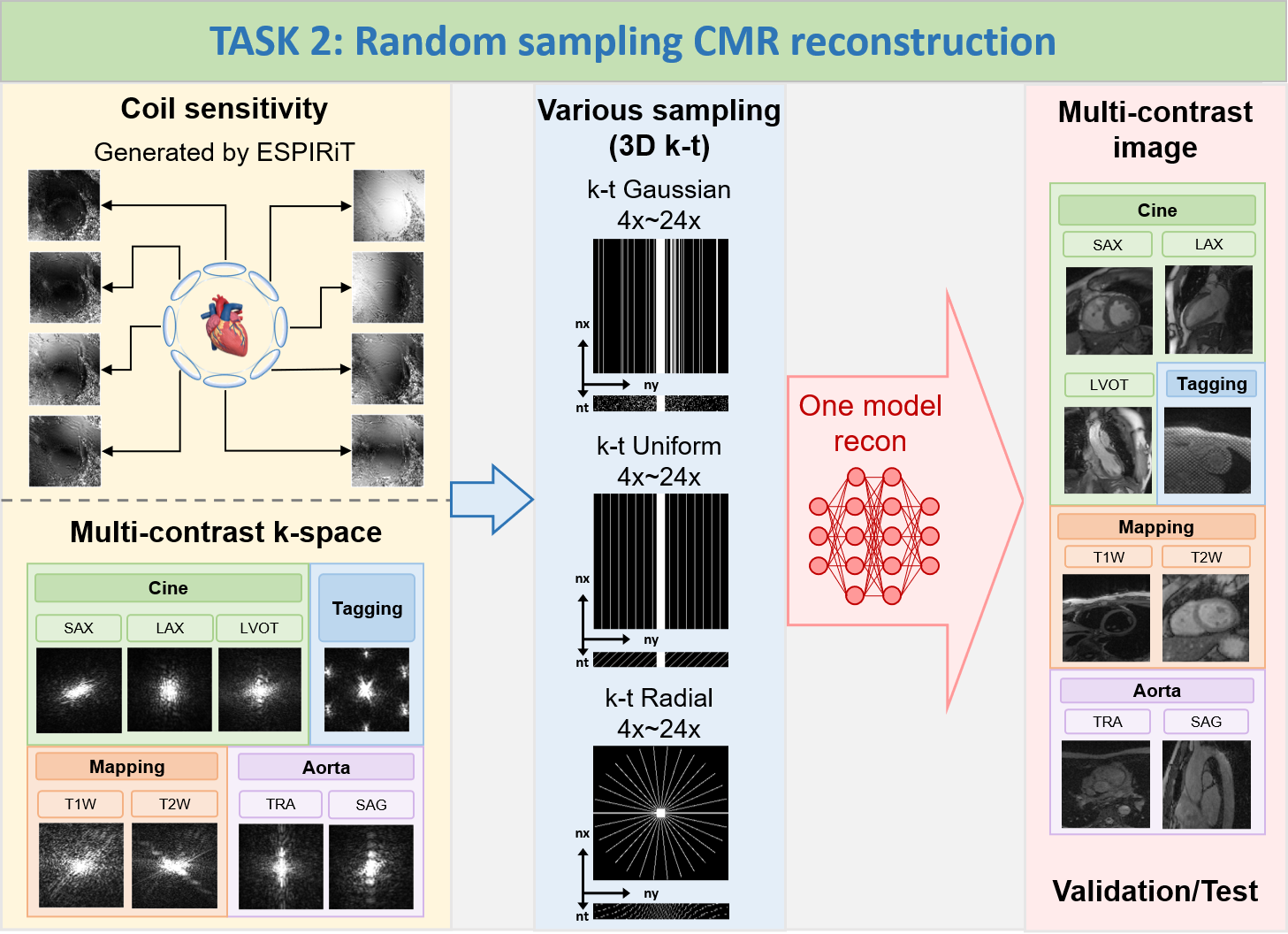
The goal of this challenge is to develop a
sampling-universal model that can robustly reconstruct CMR images 1) from different k-space trajectories
(uniform, Guassian, and pseudo radial undersampling with temporal/parametric interleaving); 2) at different
acceleration factors (acceleration factors from 4x to 24x, ACS not included for calculations). The proposed
method is supposed to leverage deep learning algorithms to exploit the potential of random sampling, enabling
faster acquisition times while maintaining high-quality image reconstructions.
Note: In TASK 2, participants are allowed
to train only one universal model to reconstruct
various data at the different undersampling scenarios (including different k-space trajectories: uniform,
Guassian, and pseudo radial undersampling with temporal/parametric interleaving; and different acceleration
factors: 4x, 8x, 12x, 16x, 20x, 24x, ACS not included for calculations); TrainingSet includes Cine, Aorta,
Mapping, and Tagging; ValidationSet and TestSet also include Cine, Aorta, Mapping, and Tagging; the
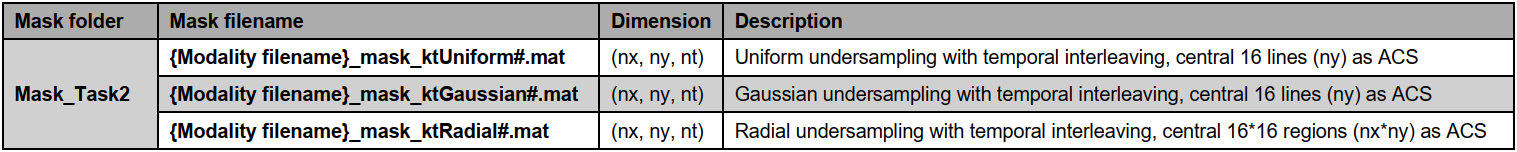
data
size of Cine, Aorta, Mapping, and Tagging is 5D (nx,ny,nc,nz,nt); the size of all undersampling masks is 3D
(nx,ny,nt), the central 16 lines (ny, in ktUniform and ktGaussian) or central 16x16 regions (nx*ny,
in
ktRadial) are always fully sampled to be used as autocalibration signals (ACS).
1) Scanner:
Siemens 3T MRI scanner (MAGNETOM Vida)
2) Image acquisition: We follow the recommendations of
CMR exams reported in the previous publication (doi: 10.1007/s43657-02100018x, 10.1007/s43657-021-00018-x).
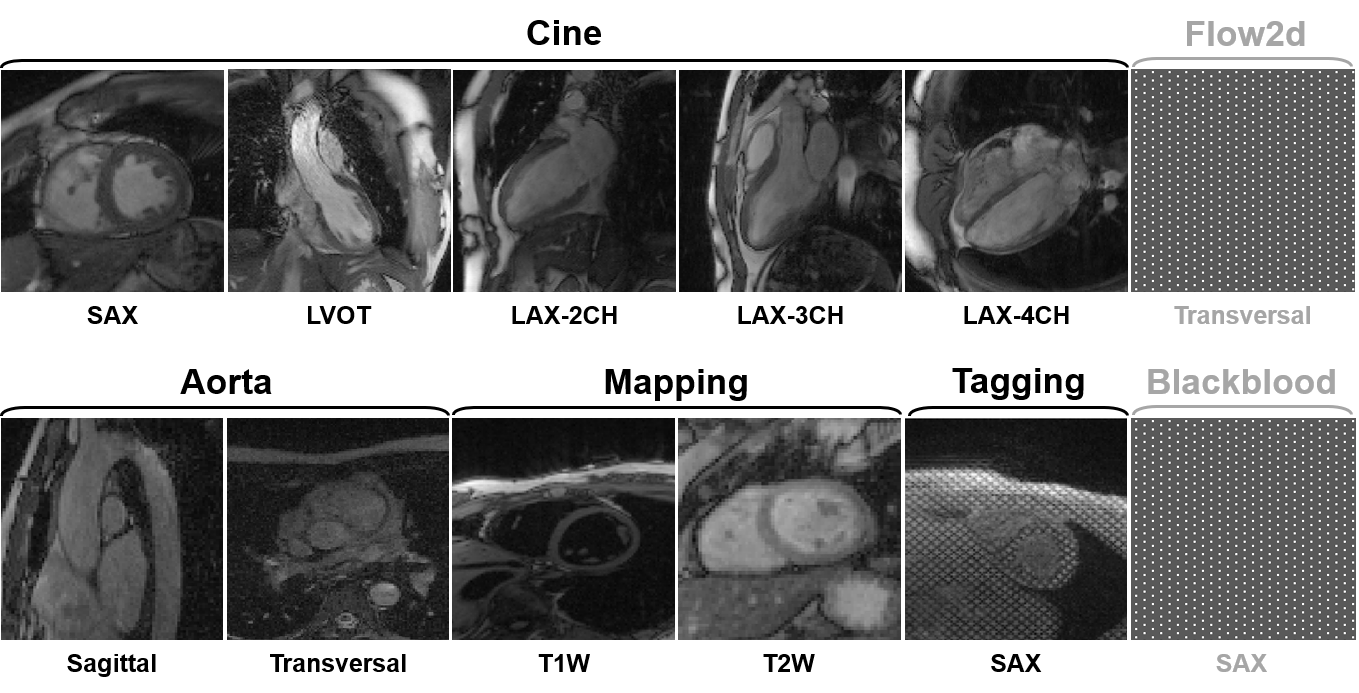
3) Dataset overview: The dataset will include multi-contrast
k-space data, consisting of cardiac cine, T1/T2 mapping, tagging, phase-contrast (i.e., flow2d), and dark-blood
imaging. It also includes imaging of different anatomical views like long-axis (LAX, including 2-chamber,
3-chamber, and 4-chamber), short-axis (SAX), left ventricul aroutflow tract (LVOT), and aortic (transversal and
sagittal views).

4) Scan protocol: We use 'TrueFISP' sequence for cine,
phase-constrast (i.e., flow2d), and tagging, and 'FLASH' sequence for T1/T2 mapping and dark-blood imaging. For
T1/T2 mapping, signals are collected at the end of the diastole with ECG triggering. Typically, 5~15 slices are
acquired for each contrast. The cardiac cycle is segmented into 12~25 phases with a temporal resolution of
around 50 ms. Typical geometrical parameters include: spatial resolution 1.5×1.5 mm2, slice thickness 8.0 mm,
and slice gap 4.0 mm.
5) Pre-processing:The raw k-space data exported from the
scanner will be processed and transformed to '.mat' format using the script provided by our vendor. A readme
file will be provided to describe the content and usage of the data.
Details of data types in Task2

Details of mask types in Task2
Metrics:
PSNR,
SSIM and NMSE
between reconstructed images and ground truth images (fully sampled data).
Ranking methods:
During the testing
and ranking phase, we will invite three radiologists to independently score the
top five teams ranked by SSIM. The scoring will cover three
aspects: image
quality, image artifacts, and clinical utility. We will consider both the
radiologists' scores and the SSIM results to generate a comprehensive ranking.
We
will calculate PSNR, SSIM and NMSE for quantitative evaluations, while we only use
SSIM as our final ranking score.
Participating
teams are required to submit docker containers and process all the cases in the
test set on our server. For the cases without valid output, we will assign it
to the lowest value of metric.
1) To ensure the fairness of this challenge, you are only allowed to use the datasets provided by fastMRI,
CMRxRecon2023, and CMRxRecon2024. Data augmentation based on the training dataset is allowed.
2) In TASK 2, participants are allowed to train only one
universal model to reconstruct various data at the aforementioned different undersampling scenarios.
created with
Website Builder Software .