





1. Sign up on the Synapse website.
2. Please fill out the registration form, including your team name, team members, Synaspe account, and sign the data agreement.
3. Send both the registration form and the data agreement to CMRxRecon@outlook.com.
Email Format
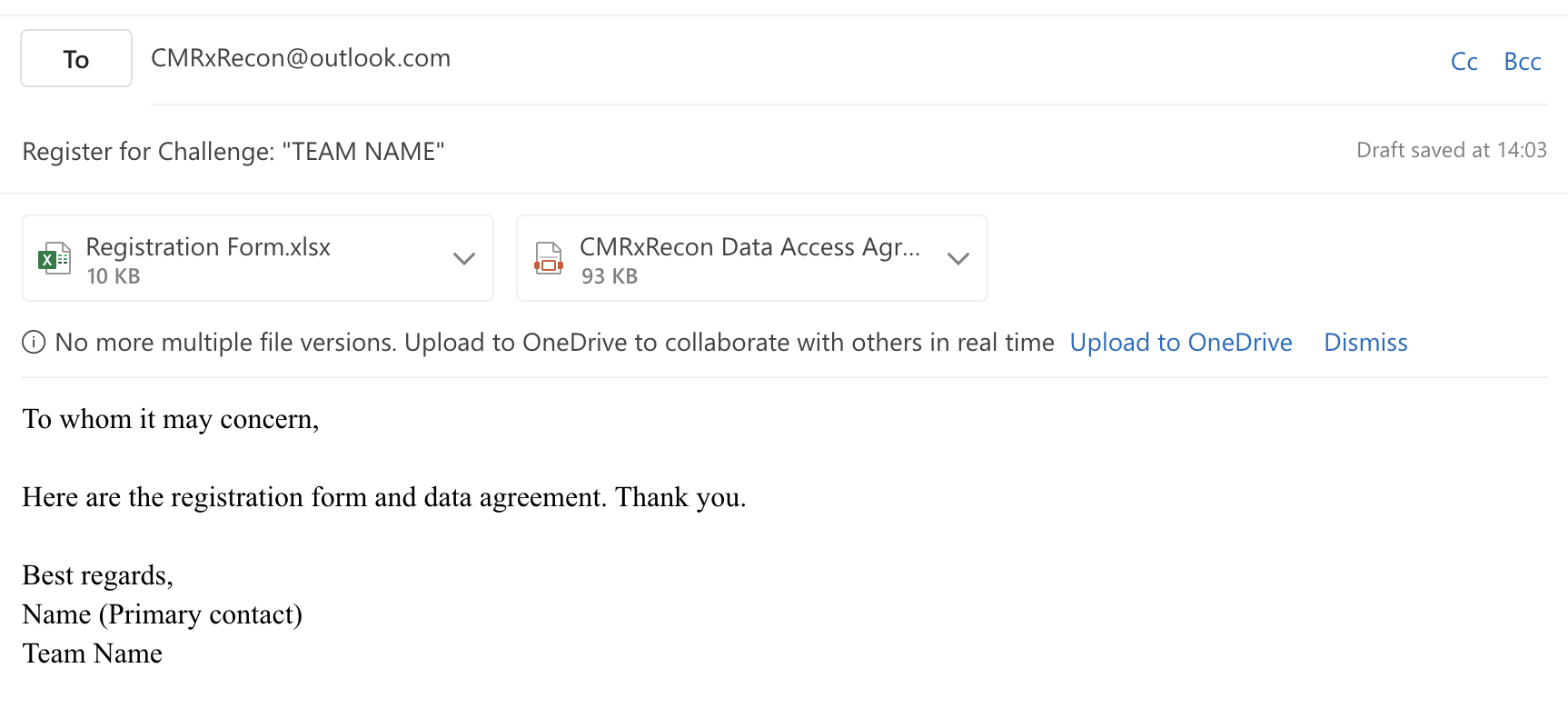
Note: Please use lowercase letters and numbers in your team name. Do not include any spacing or special characters.
Correspondence will not be directed to the personal email address of the organizers, unless a response has not been received in four business days.
Once we received the registration form and the data agreement from your team, you have the access to download the data.
[10 May, 2023] Training data and validation data will be released. The results can be evaluated from 30 May, 2023.
Please follow the instructions on Synapse to download the data.
We will send you the link and password to download the data (via
Google Drive
, One Drive, and
Baidu Netdisk)
We provide the code to facilitate the use of the data we release at https://github.com/CmrxRecon/CMRxRecon.
A brief description of the provided package is as follows:
a) 'CMRxReconDemo': contains parallel imaging reconstruction code;
b) 'ChallengeDataFormat': explain the challenge data and the rules for data submission;
c) 'Evaluation': contains image quality evaluation code for validation and testing;
d) 'Mapping': contains fitting code for T1 mapping and T2 mapping;
e) ‘Download_Dataset_Check’: check whether the dataset is completely and rightly downloaded;
f) 'Submission': contains the structure for challenge submission.
Validation of the received docker on unseen test set will be performed on a cloud server with a configuration as follows:
You are free to use and/or refer to the CMRxRecon challenge and datasets in your own research after the embargo period (Dec 2023), provided that you cite the following manuscripts:
Reference of the imaging acquisition protocol:
1. Wang C, Li Y, Lv J, et al. Recommendation for Cardiac Magnetic Resonance Imaging-Based Phenotypic Study: Imaging Part. Phenomics. 2021, 1(4): 151-170. https://doi.org/10.1007/s43657-021-00018-x
Other reference (optional for citation):
1. Wang C, Jang J, Neisius U, et al. Black blood myocardial T2 mapping. Magnetic resonance in medicine. 2019, 81(1): 153-166. https://doi.org/10.1002/mrm.27360
2. Lyu J, Li G, Wang C, et al. Region-focused multi-view transformer-based generative adversarial network for cardiac cine MRI reconstruction[J]. Medical Image Analysis, 2023: 102760. https://doi.org/10.1016/j.media.2023.102760
3. Qin C, Schlemper J, Caballero J, et al. Convolutional recurrent neural networks for dynamic MR image reconstruction. IEEE transactions on medical imaging, 2018, 38(1): 280-290. https://doi.org/10.1109/TMI.2018.2863670.
4. Qin C, Duan J, Hammernik K, et al. Complementary time‐frequency domain networks for dynamic parallel MR image reconstruction. Magnetic Resonance in Medicine, 2021, 86(6): 3274-3291. https://doi.org/10.1002/mrm.28917
5. Lyu J, Tong X, Wang C. Parallel Imaging With a Combination of SENSE and Generative Adversarial Networks (GAN). Quantitative Imaging in Medicine and Surgery. 2020, 10(12): 2260–2273. https://doi.org/10.21037/qims-20-518.
6. Lyu J, Sui B, Wang C, et al. DuDoCAF: Dual-Domain Cross-Attention Fusion with Recurrent Transformer for Fast Multi-contrast MR Imaging. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, 2022: 474-484.
7. Wang S, Qin C, Wang C, et al. The Extreme Cardiac MRI Analysis Challenge under Respiratory Motion (CMRxMotion). arXiv preprint arXiv:2210.06385, 2022.
We will provide monetary awards for the top 3 winners of each task. The prize pool is exclusively sponsored by Siemens.
Task 1. Accelerated cine construction
Task 2. Accelerated T1/T2 mapping
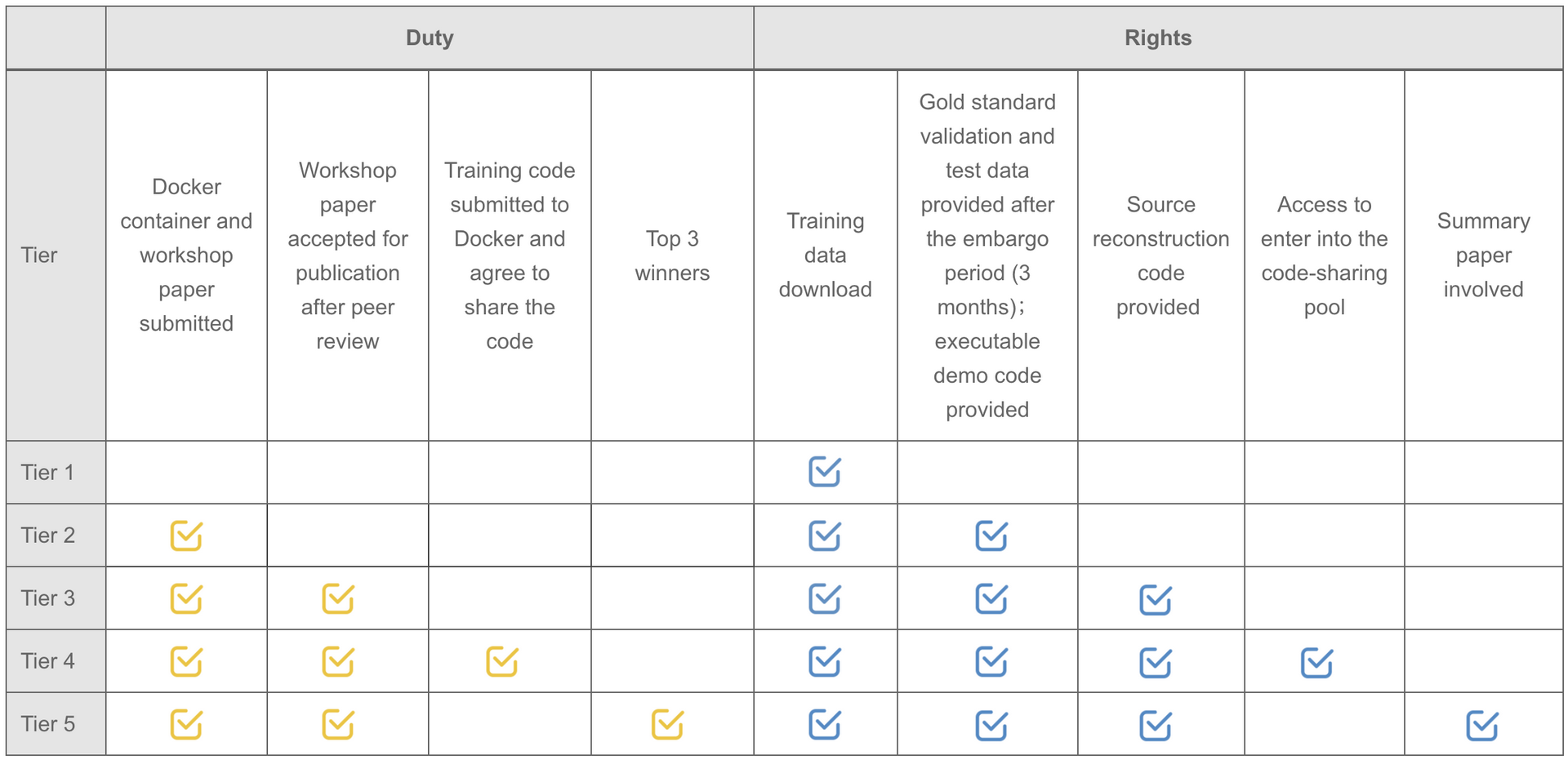
Note: Participants are not required to upload the complete training code. But teams willing to upload the original training code will be automatically entered into the code-sharing pool.
created with
Website Builder Software .