





Welcome to the Cardiac MRI
Reconstruction Challenge (CMRxRecon)!
The
CMRxRecon Challenge is a part of the 26th International Conference on Medical Image Computing and
Computer Assisted
Intervention, MICCAI 2023, which will be held from October 8th to 12th 2023 in Vancouver
Convention Centre Canada.
Cardiac magnetic resonance imaging (CMR)
has become an important imaging modality for diagnosing cardiac diseases due to its superior soft tissue
contrast and non-invasiveness. However, an inherent drawback of MRI is that the imaging speed is
particularly slow, which will cause discomfort to patients and introduce motion artifacts into
images. CMR image reconstruction from highly under-sampled k-space (raw data) has become a hot topic
in recent years.
So far, a large number of AI-based image reconstruction algorithms have shown the
potential to improve imaging performance through increasing the data under-sampling factor. However, the
field of CMR reconstruction still lacks public, standardized, and high-quality datasets. To date, NYU
Langone Health has released 'fastMRI' dataset, containing multi-channel knee and brain MRI raw data. However,
these images are inadequate for 3D+1D (time domain) applications in cardiac imaging. The goal of establishing
the 'CMRxRecon' dataset is to provide a platform that enables the broad research community to participate in
this important work.
The ‘CMRxRecon’ challenge includes two
independent tasks. Each team can choose to participate one of them or both:
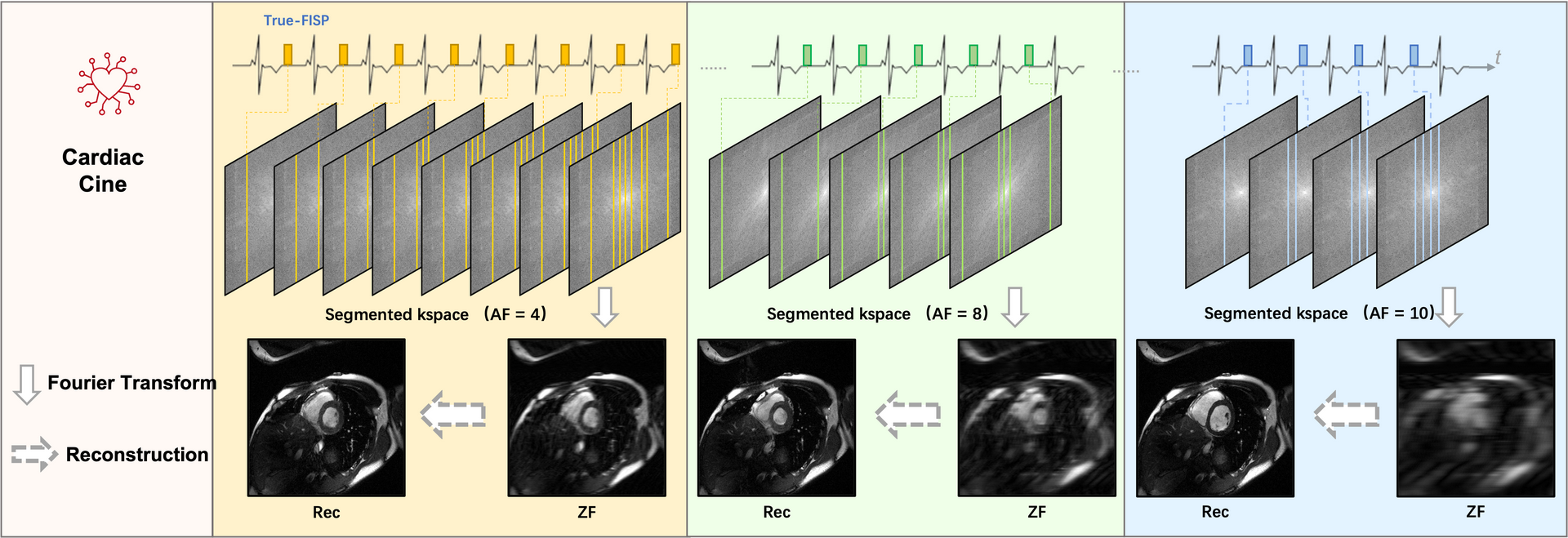
1) Accelerated
cine reconstruction
The aim of task 1 is to accelerate cine
imaging by rawdata under-sampling and address the image degradation due to motions caused by voluntary breath-holds or
cardiac arrhythmia. The final goal will be real-time cine imaging.
Task 1. Accelerated Cine Reconstruction
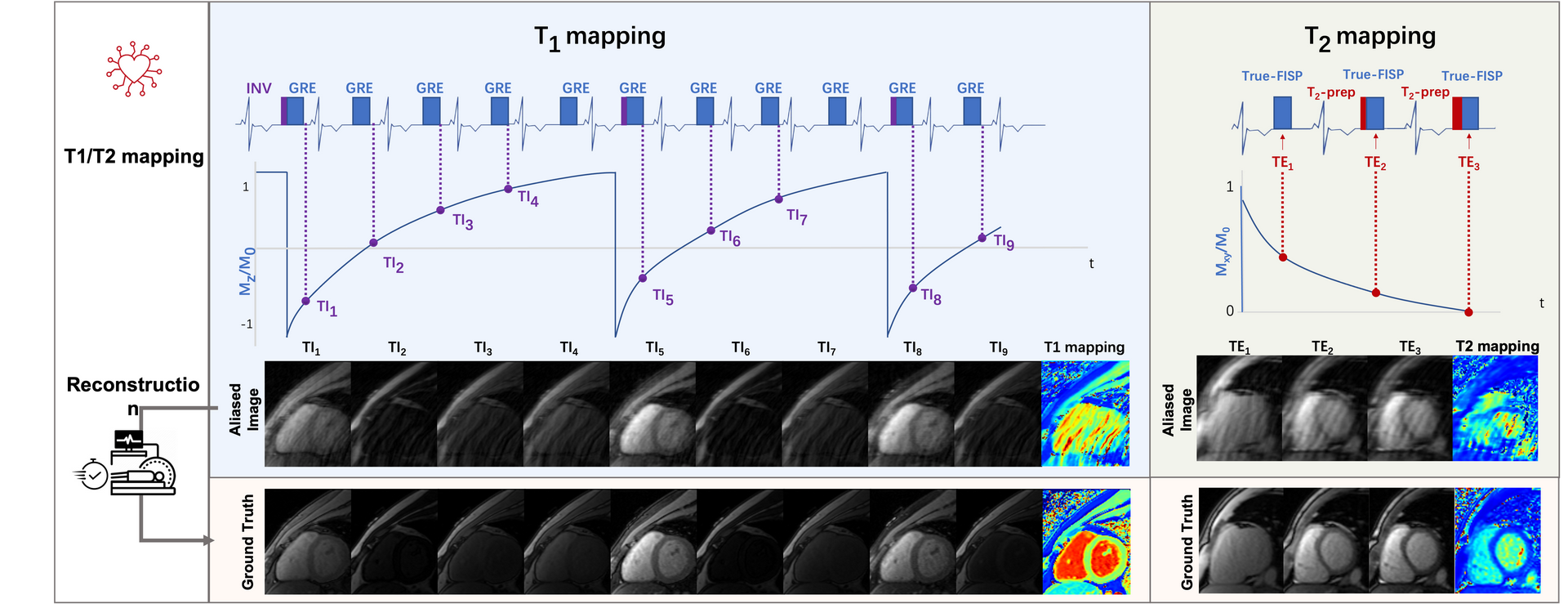
2) Accelerated
T1/T2 mapping
The aim of task 2 is to improve the T1 and T2 mapping estimation accuracy by
rawdata under-sampling and address the image degradation due to motions and under-sampled
reconstructions.
Task 2. Accelerated T1/T2 Mapping
We will provide monetary awards for the top 3 winners of each task. The prize pool is exclusively sponsored by Siemens.
Task 1. Accelerated cine construction
Task 2. Accelerated T1/T2 mapping
A total of 300 healthy volunteers from a single center were included in
this study.
The released dataset includes 120 training data, 60 validation data and 120 test
data.
Training data including fully sampled k-space data, auto-calibration lines (ACS, 24 lines) and
reconstructed images in MATLAB .mat format will be provided.
Validation data include under-sampled k-space
data with acceleration factors of 4, 8 and 10, sampling mask, and auto-calibration lines (ACS, 24 lines) will be
provided. We will withhold the ground truth images of the validation set.
Test data include under-sampled
k-space data with acceleration factors of 4, 8 and 10, sampling mask, auto-calibration lines (ACS, 24 lines) and
reconstructed images. The test data will not be available to the participants.
Scanner: Siemens 3T MRI scanner (MAGNETOM Vida).
Image acquisition: We followed the recommendations of CMR
imaging reported in the previous publication (doi:
10.1007/s43657-02100018x,
10.1007/s43657-021-00018-x[w.c.y.1]
).
1) Cine: The ‘TrueFISP’ readout was used for CINE acquisition. The collected
images include short-axis (SA), two-chamber (2CH), three-chamber (3CH) and four-chamber(4CH) long-axis (LA)
views. Typically 5~10 slices were acquired for SA cine, while a single slice was acquired for the
other views. The cardiac cycle was segmented into 12–25 phases with a temporal resolution 50 ms. For this
challenge, we provided raw k-space data of both SA (multi-slices) and LA
(multi-views).
2) Mapping: T1 mapping was conducted using a modified look-locker
inversion recovery (MOLLI) sequence, which acquired nine images with different T1 weightings (using the
4-(1)-3-(1)-2 scheme). T1 mapping was performed in SA view only. Signals were collected at the end of the
diastole with ECG triggering. T2 mapping was performed using T2-prepared (T2prep)-FLASH sequence with three
T2 weightings in SA view, with identical geometrical parameters as used in T1 mapping.
Pre-processing: The raw k-space data exported from the scanner will be pre-processed and transformed to .mat format (MATLAB). The data were compressed to 10 virtual coils (Zhang et. al MRM 2013;69(2):571-82.) for standardization and to save storages. The image quality is highly consistent before and after coil compression. The partial Fourier data were filled up using POCS algorithm (provided by michael.voelker@mr-bavaria.de). We will provide a README file that describes the content of the data and how to use it.
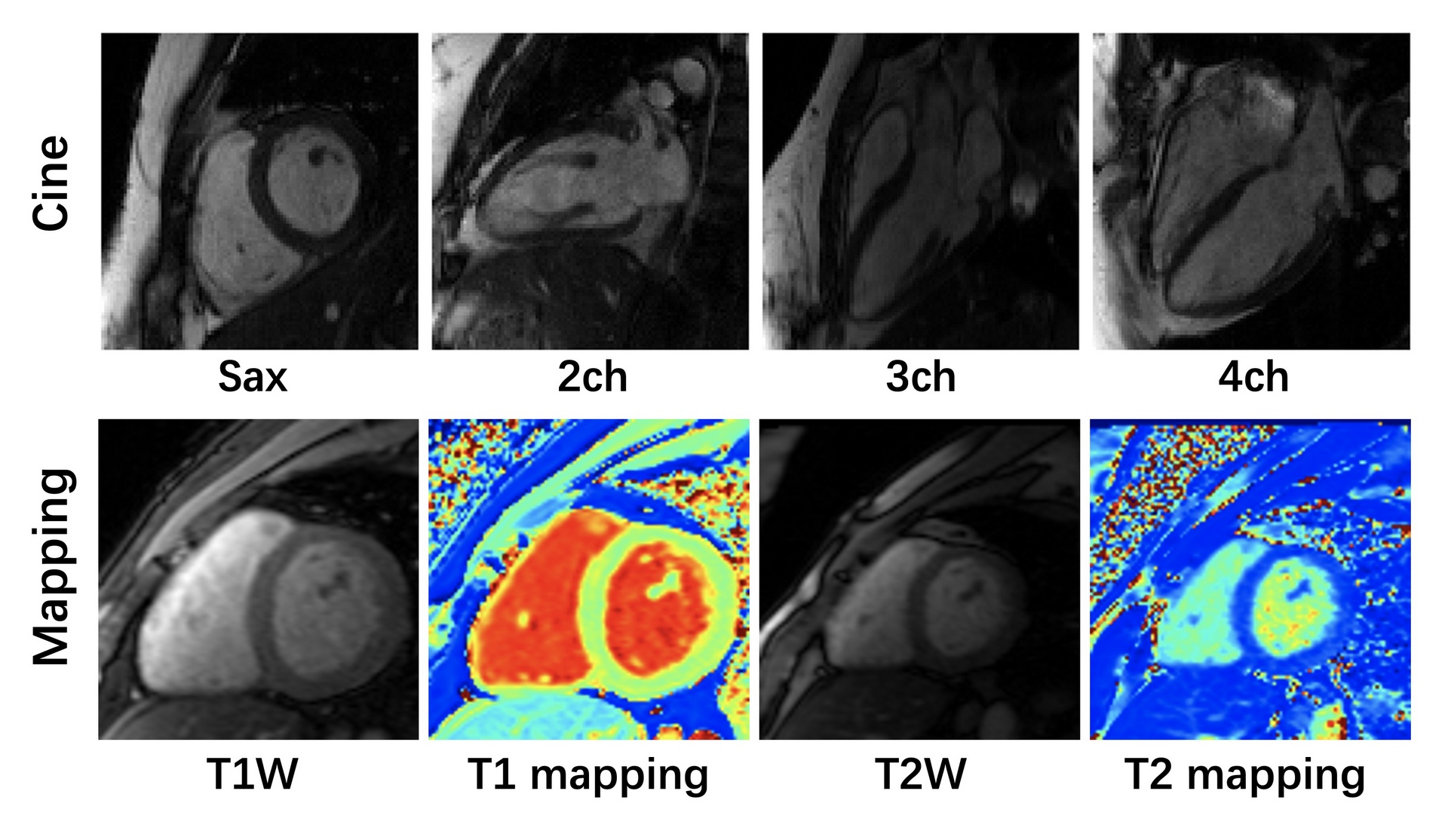
Representative Images with Manually Annotations
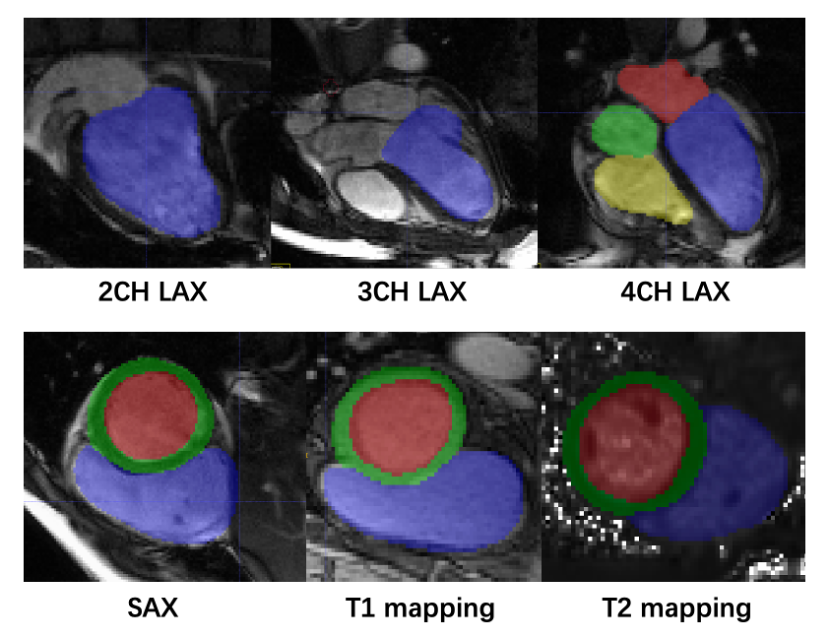
Manual segmentations of myocardium and chambers were performed by an
experienced radiologist (with more than 5 years of cardiac imaging experience) using ITK-SNAP (version 3.8.0).
The segmentation labels and the corresponding images were stored in NIFTI format, maintaining the original image
coordinates.
For the LAX cine images, four cardiac chambers have been labeled as follows:
a) Left atrium
- label 1;
b) Right atrium - label 2;
c) Left ventricle - label 3;
d) Right ventricle - label 4.
For the SAX cine images, we performed the following labeling:
a) Left ventricle - label 1;
b) Left
ventricular myocardium - label 2;
c) Right ventricle - label 3.
The annotations of both T1 mapping and
T2 mapping were the same as SAX cine.
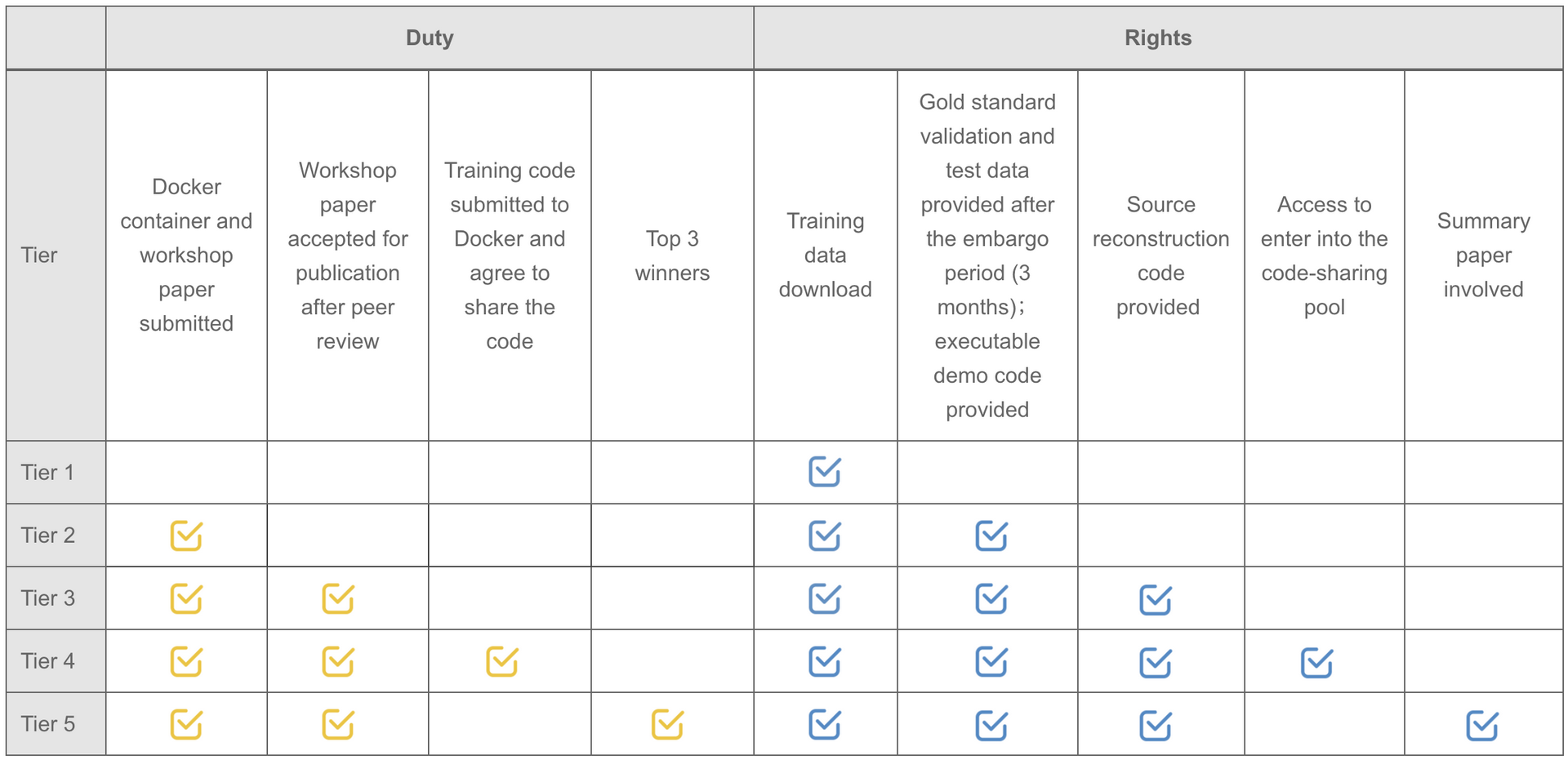
Note: Participants are not required to upload the complete training code. But teams willing to upload the original training code will be automatically entered into the code-sharing pool.
The schedule of the challenge is as follows. All deadlines are Pacific Standard Time (PST +0:00).
created with
Website Builder Software .