



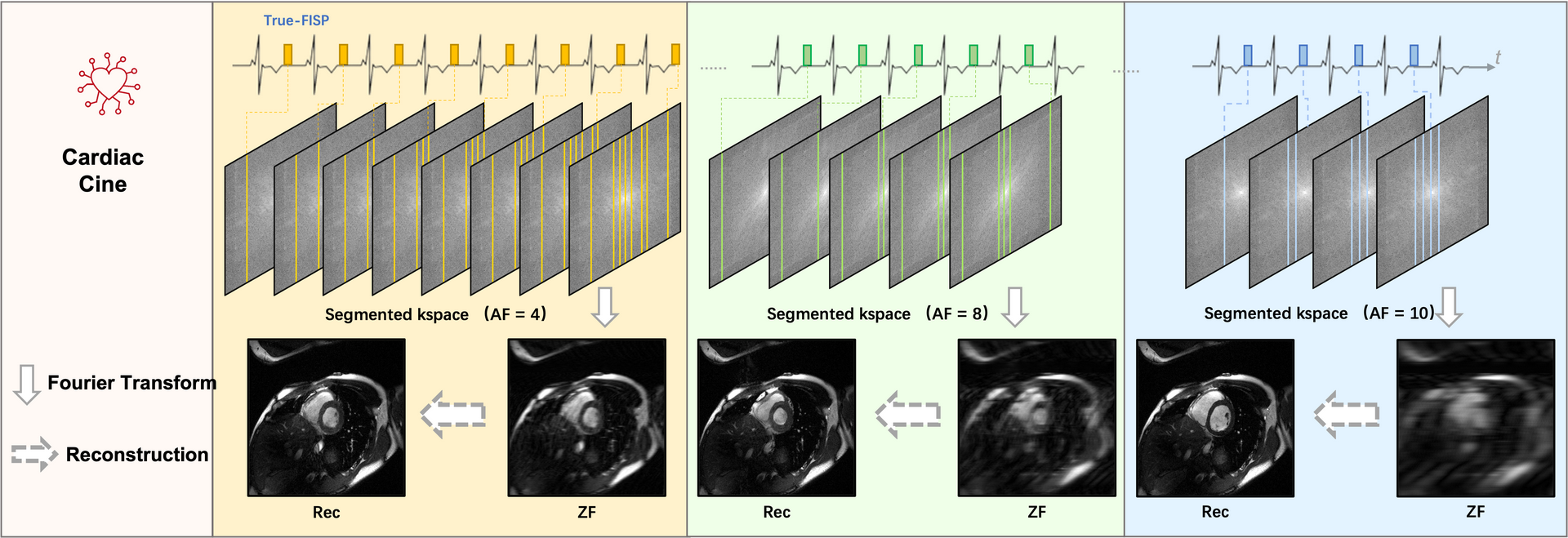
Cine is the current gold standard for the noninvasive evaluation of cardiac functional parameters. Typically, cine MRI is obtained by electrocardiography (ECG) -gated segmented sequence since the full k-space cannot be obtained within one acquisition window, where the segments of the entire k-space are read out over multiple cardiac cycles. For patients with voluntary breath-holds or cardiac arrhythmia, image degradation will occur during the long acquisition time and motions, thus affecting the further diagnosis. Accelerated cine MRI has the capability to address these limitations, in which the k-space of all temporal phases can be filled up in a fewer cardiac cycles and in a single breath-hold. It is also possible to perform “real-time” imaging by further accelerated reconstructions, therefore artifacts associated with respiratory motion and arrhythmia can be largely reduced. This challenge aims to establish a platform for fast cine reconstruction and provide a benchmark multi-view dataset that enables the broad research community to promote advances in this area of research.
Scanner: Siemens 3T MRI scanner (MAGNETOM Vida).
Image acquisition: We followed the recommendations of CMR imaging reported in the previous publication ( d oi: 10.1007/s43657-02100018x, 10.1007/s43657-021-00018-x [w.c.y.1] ). The ‘TrueFISP’ readout was used for CINE acquisition. The collected images include short-axis (SA), two-chamber (2CH), three-chamber (3CH) and four-chamber(4CH) long-axis (LA) views. Typically 5~10 slices were acquired for SA cine, while a single slice was acquired for the other views. The cardiac cycle was segmented into 12–25 phases with a temporal resolution 50 ms. For this challenge, we provided raw k-space data of both SA (multi-slices) and LA (multi-views). Typical scan parameters: spatial resolution 2.0×2.0 mm2, slice thickness 8.0 mm, and slice gap 4.0 mm.
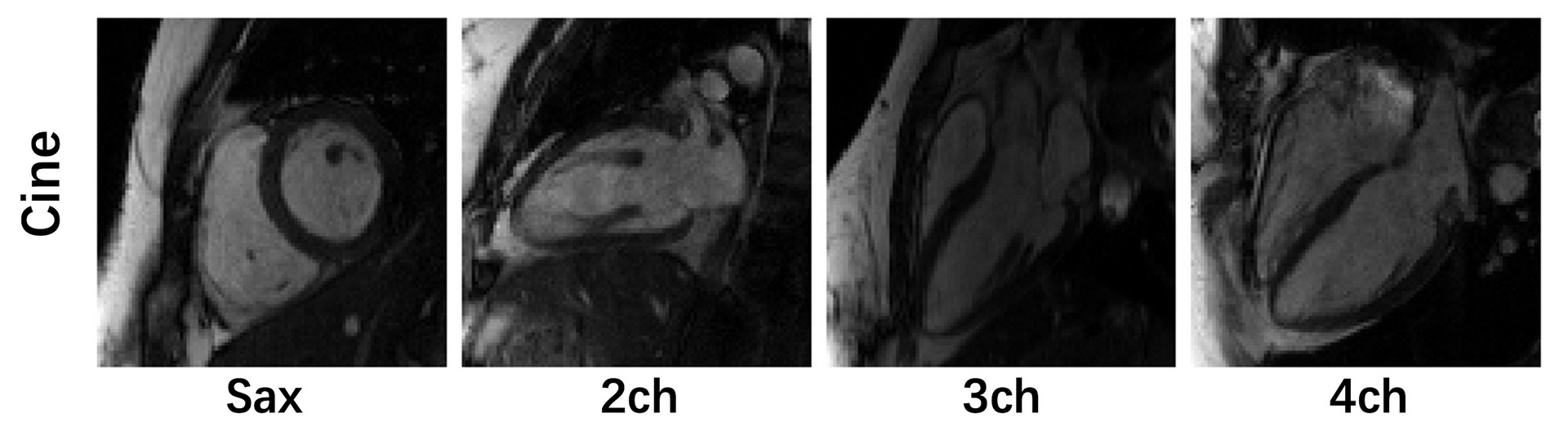
Pre-processing: The raw k-space data exported from the scanner will be pre-processed and transformed to .mat format (MATLAB). The data were compressed to 10 virtual coils (Zhang et. al MRM 2013;69(2):571-82.) for standardization and to save storages. The image quality is highly consistent before and after coil compression. The partial Fourier data was filled up using POCS algorithm (provided by michael.voelker@mr-bavaria.de). We will provide a README file that describes the content of the data and how to use it. Taking Long axis (Lax) cine as an example, the format of data is as follows:
Details of the Data Types of Cardiac Cine
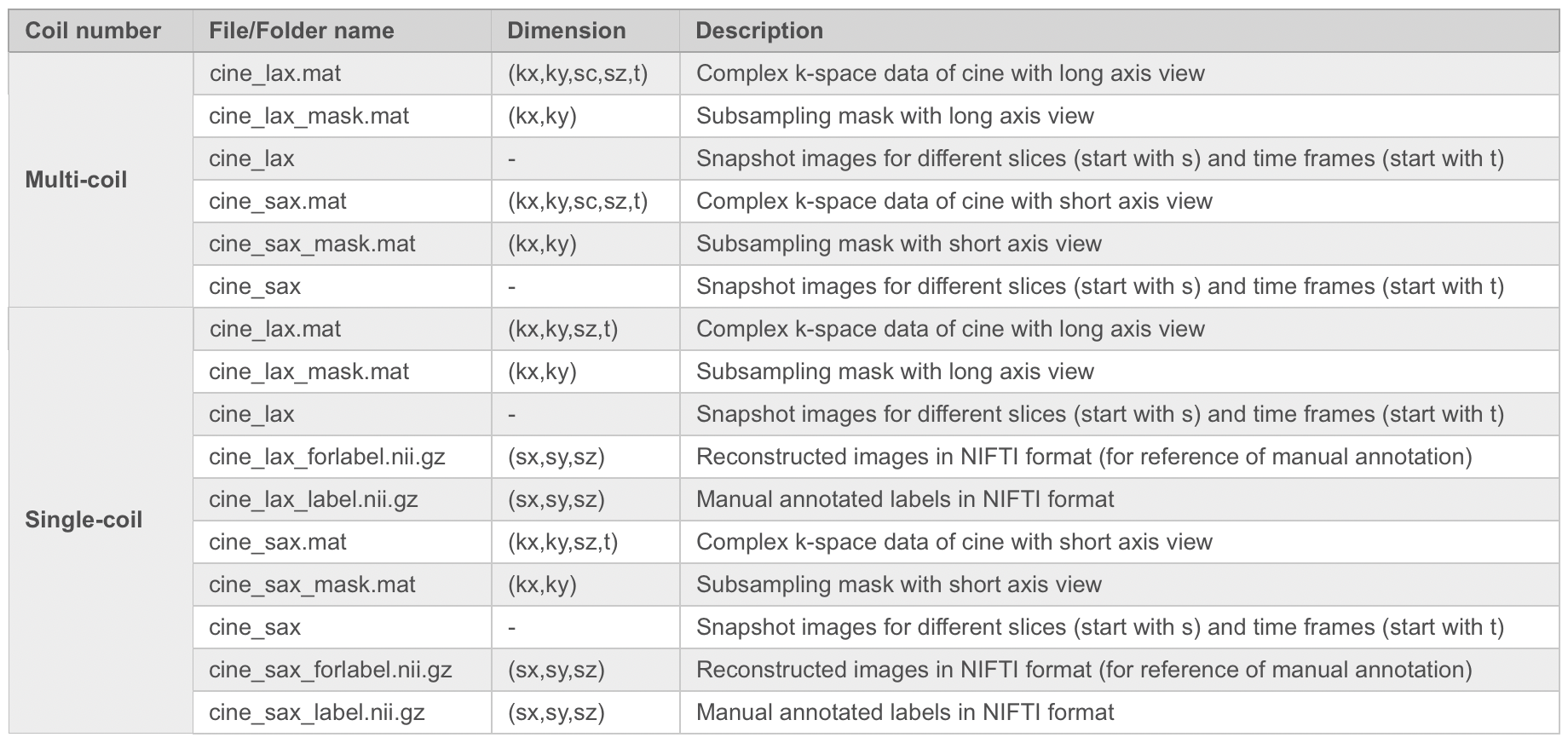
Note: kx: matrix size in x-axis (k-space); ky: matrix size in y-axis (k-space); sc: coil array number (compressed to 10); sx: matrix size in x-axis (image); sy: matrix size in y-axis (image); sz: slice number for short axis view, or slice group for long axis (i.e., 3ch, 2ch and 4ch views); t: time frame.
1. Multi-coil data:
Data: cine_lax.mat
# cine with long axis view (including 3ch, 2ch and 4ch views within the sz dimension).
# variable name:
# "kspace_full" for full kspace
# "kspace_sub04" for subsampling factor of 4 along ky
# "kspace_sub08" for subsampling factor of 8 along ky
# "kspace_sub10" for subsampling factor of 10 along ky
# data type: complex kspace data with the dimensions (kx,ky,kc,kz,t), the central 24 lines (ky) are always full sampled to be used as calibration lines
-kx: matrix size in x-axis (kspace)
-ky: matrix size in y-axis (kspace)
-sc: coil array number (compressed to 10)
-sz: slice number (short axis view); slice group (long axis, i.e., 3ch, 2ch and 4ch views)
-t: time frame
Data: cine_lax_mask.mat
# subsampling mask with long axis view (including 3ch, 2ch and 4ch views within the sz dimension), the mask is fixed among different sc, sz and t.
# variable name:
# "mask04" for subsampling factor of 4 along ky
# "mask08" for subsampling factor of 8 along ky
# "mask10" for subsampling factor of 10 along ky
# data type: binary data with the dimensions (kx,ky), the central 24 lines (ky) are always full sampled to be used as calibration lines
-kx: matrix size in x-axis (kspace)
-ky: matrix size in y-axis (kspace)
2. Single-coil data:
Data: cine_lax.mat
# cine with long axis view (including 3ch, 2ch and 4ch views within the sz dimension).
# variable name:
# "kspace_single_full" for full kspace
# "kspace_single_sub04" for subsampling factor of 4 along ky
# "kspace_single_sub08" for subsampling factor of 8 along ky
# "kspace_single_sub10" for subsampling factor of 10 along ky
# data type: complex kspace data with the dimensions (kx,ky,kz,t), the central 24 lines (ky) are always full sampled to be used as calibration lines
-kx: matrix size in x-axis (kspace)
-ky: matrix size in y-axis (kspace)
-sz: slice number (short axis view); slice group (long axis, i.e., 3ch, 2ch and 4ch views)
-t: time frame
Data: cine_lax_mask.mat
# subsampling mask with long axis view (including 3ch, 2ch and 4ch views within the sz dimension), the mask is fixed among different sc, sz and t.
# variable name:
# "mask04" for subsampling factor of 4 along ky
# "mask08" for subsampling factor of 8 along ky
# "mask10" for subsampling factor of 10 along ky
# data type: binary data with the dimensions (kx,ky), the central 24 lines (ky) are always full sampled to be used as calibration lines
-kx: matrix size in x-axis (kspace)
-ky: matrix size in y-axis (kspace)
Data: cine_lax_forlabel.nii.gz
# reconstructed images in nifti format (for reference of manual annotation)
-sx: matrix size in x-axis
-sy: matrix size in y-axis
-sz: slice number (short axis view); slice group (long axis view)
Data: cine_lax_label.nii.gz
# manual annotated labels in nifti format
# label 1 for left ventricle
# label 2 for myocardium
# label 3 for right ventricle
-sx: matrix size in x-axis
-sy: matrix size in y-axis
-sz: slice number (short axis view); slice group (long axis view)
Radiologists' scoring according to the Likert score system will be performed on the top 5 participant groups in regard to the images' overall the quality, artifacts, sharpness and contrast-to-noise ratio using the ground truth as the reference.
Likert Score
| 4-point score | Grade |
| 1 | Excellent |
| 2 | More than adequate for diagnosis |
| 3 | Adequate for diagnosis |
| 4 | Non diagnostic |
The submission instructions will be released on the Synapse platform.
created with
Website Builder Software .